Research Services:
Whole gene expression microarray analysis (CLARIOM S) – HUMAN / MOUSE / RAT
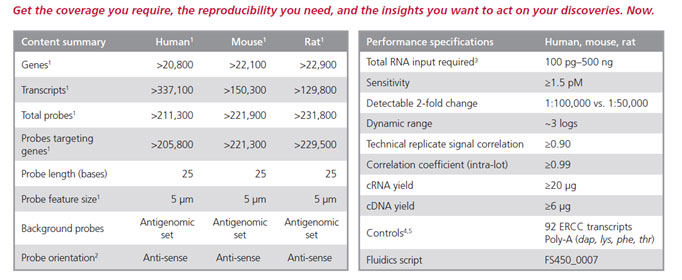
Obtain a gene-level view of the human transcriptome with the Clariom S Assays for human. Based on industry-leading microarray technology, the novel Human Clariom S Assay design provides extensive coverage of all known well-annotated genes, compatibility with clinical sample types, scalable formats, and flexible data analysis software. Clariom S Assays are the tools of choice to find expression biomarkers with known function as quickly, easily, and cost-effectively as possible.
Key features:
Robust results even from precious samples
- Generate expression profiles from as little as 100 pg oftotal RNA—as few as 10 cells
- Analyze RNA from a wide variety of sample types,including cells, whole blood, and fresh/fresh-frozen orformalin-fixed, paraffin-embedded (FFPE) tissues
- Preserve sample integrity and reduce variability with noneed for globin or ribosomal RNA removal
Whole Transcriptome Gene expression analysis (CLARIOM D) – HUMAN / MOUSE / RAT
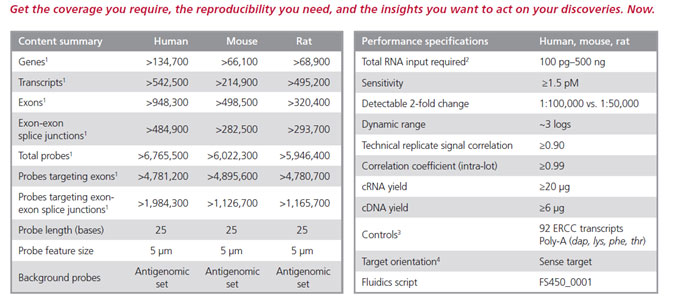
Accelerate your biomarker discovery with Clariom D solutions—the next generation of transcriptome-level profiling tools—providing a highly detailed view of the transcriptome for the fastest path to actionable results. Available for human, mouse, and rat, Clariom D solutions allow translational scientists to generate high-fidelity biomarker signatures quickly and easily with a design that provides the most intricate transcriptome-wide gene- and exon-level expression profiles including the ability to detect alternative splicing events of coding and long non-coding RNA, in a single three-day experiment.
Get all the data you need. Now.
- Rapidly identify complex disease signatures from as many as 540,000 transcripts, ensuring biomarkers are not missed.
- Confidently detect genes, exons, and alternative splicing events that give rise to coding and long non-coding RNA isoforms.
- Detect rare and low-expressing transcripts otherwise missed by common sequencing practices.
When you have precious samples, get it right the first time.
- Generate robust expression profiles from as little as 100 pg of total RNA—as few as 10 cells.
- Use RNA from various sample types including whole blood, cultured cells, and fresh/fresh frozen or FFPE tissues.
- Preserve sample integrity and reduce data variability with an assay that does not require a globin or rRNA removal step.
OncoScan™ FFPE Assay
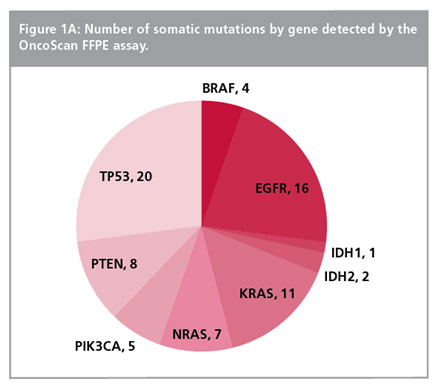
Copy number analysis in solid tumors is rapidly gaining importance in cancer diagnosis, prognosis, and therapy selection1. OncoScan™ FFPE Assay Kit is capable of determining highly accurate copy number changes and allelic imbalances, including loss of heterozygosity (LOH) in solid tumors from limited amounts of highly modified and degraded FFPE-derived DNA.
Key Features:
- Low sample input, fast results − from only 80 ng of FFPE-derived DNA to results in 48 hours
- One assay, multiple data views – whole-genome copy number, loss of heterozygosity (LOH) at ≤10 Mb across the genome, and detection of key somatic mutations
- Detection of copy number changes found in as low as 25% of aberrant cells with a linear copy number range of 50+ copies
- High-resolution (50–125 kb) copy number detection in priority cancer genes
- Rapid analysis – visualization of copy number data for hundreds of samples in minutes
- Detection of frequently tested somatic mutations at ~20% sensitivity
NEXT GENERATION SEQUENCING SERVICES:
Whole Genome Sequencing
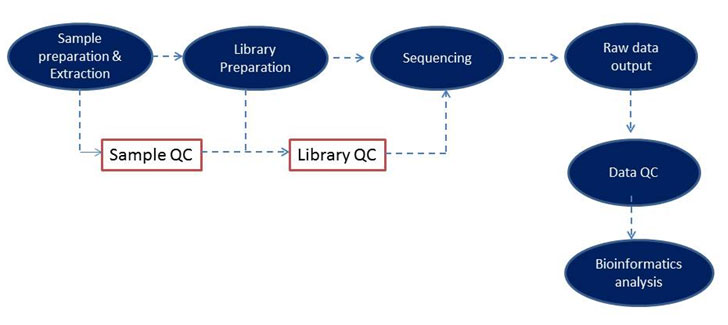
3i Molecular Solutions offer an end-to-end solution for next-generation sequencing as a service for a wide range of sample types (DNA, RNA as shown in Figure 1). To learn more about our services click on the tabs in the workflow below.
Whole Exome Sequencing:
Whole Exome Sequencing using NGS have been broadlyacknowledged to speed up and lessen the cost of sequencing genomes for basic research as well as use of genomic data for a wide range of applications: Genome wide association studies for complex diseases, variant calling to identify clinically actionable mutations, single nucleotide polymorphism and INDELS. At 3i Molecular Solutions, we regularly perform Whole Genome Sequencing and Exome Sequencing and analyses using best practices.
Whole Transcriptomics Sequencing
NGS enables gene expression analysis from normal and disease tissues with high-sensitivity. Profiling of coding and non-coding RNAs is important in revealing molecular mechanisms of development and disease. With the several options that exist for gene expression profiling, it is also promising to perform whole transcriptome profiling ranging from a single-cell to thousands of cells in a sample. Additionally, current methodologies allow for profiling of transcriptomes from intact RNA and RNA degraded due to the storage and isolation conditions (such as FFPE processed tissue blocks, cell-free nucleic acids etc). At 3i Molecular Solutions, we offer solutions, for performing gene expression analysis from a wide-range of sample types.

MICROARRAY DATA ANALYSIS
- Ø CNV data analysis using ChAS
- Ø Gene expression data analysis using R and TAC
- Ø Quality control & Normalization
- Ø Differential Expressed Genes (Upregulated &Downregulated), Different plots (Heatmap, volcano plot etc) using R
- Ø Protein-Protein interactions and pathway analysis
NGS DATA ANALYSIS
- Ø Data Retrieval (NCBI SRA) & Introduction to data types
- Ø Read Quality Check and trimming (FastQC&Cutadapt)
- Ø Alignment of reads using reference Genome SNP and indel Calling SNP annotations
- Ø Gene Expression Quantification (Coverage, FPKM), Differential expression analysis (Cufflink, cuffmerge&cuffdiff)
- Ø Pathway & Gene ontology enrichment analysis
- Ø Pathway Network Analysis

